rctvolume.py in appion 3.0
Added by Jason van Rooyen almost 11 years ago
I am trying to follow the RCT workflow in Appion.
Having used template picking to pick particles from tilt pairs, I then inspected and corrected pics using “Align and edit tilt pairs”. I then extracted particles into tilted and untilted stacks. They have a similar number of images and upon inspection I can confirm that they are indeed the correct pairs. I then aligned the zero-tilt image and classified.
However, when using these class averages to generate a RCT volume, I get the following error:
running:
/usr/local/bin/appion/rctVolume.py --projectid=7 --description="test" --rundir=/myami_data/MYAMI/APPION/14sep25b/rctvolume/rct9clust54class32104 --runname=rct9clust54class32104 --cluster-id=54 --classnums=3,2,1,0,4 --tilt-stack=43 --mask-rad=24 --num-iters=3 --zoom=0.9 --mass=960 --lowpassvol=10 --highpasspart=800 --median=3 --commit --expid=57 --jobtype=rctvolume
gives:
In file "/usr/local/bin/appion/rctVolume.py", line 357, in makeEulerDoc:
totrot = -1.0*(notrot + inplane)
TypeError: unsupported operand type(s) for +: 'float' and 'NoneType'
Following the program with the debugger PUDB, it seems that the the “inplane” and “mirror” values returned by
getParticleInPlaneRotation(self, tiltstackpartdata) on line 149 are both equal to “none”.
Is there a way to check the parameters associated with tilt-pair stacks to see if the parameters exist in the database?
Replies (8)
RE: rctvolume.py in appion 3.0 - Added by Anchi Cheng almost 11 years ago
We have not had this problem before, so debug will be harder.
Your debugger result does point to no value uploaded during alignment. Which alignment method did you use ?
Any way, the easiest way to look at the database is to use phpMyAdmin or equivalent.
You will need to navigate in there to get to database for project with id of 7, probably called "ap7".
1. Find in ApClusteringStackData the row of data with `DEF_id` of 54, and then get the id value under the column `REF|ApAlignStackData|alignstack`
2. Find in ApAlignParticleData, search for the id you got from step 1 under the column `REF|ApAlignStackData|alignstack`
The result will be parameters uploaded for that alignment stack.
RE: rctvolume.py in appion 3.0 - Added by Neil Voss almost 11 years ago
What program/method did you use for the alignment of the untilted particles?
RE: rctvolume.py in appion 3.0 - Added by Jason van Rooyen almost 11 years ago
I tried both EMAN refine2d and Xmipp max-liklihood.
However, the entire `ApAlignParticleData` table is NULL.
RE: rctvolume.py in appion 3.0 - Added by Jason van Rooyen almost 11 years ago
This is what I found: 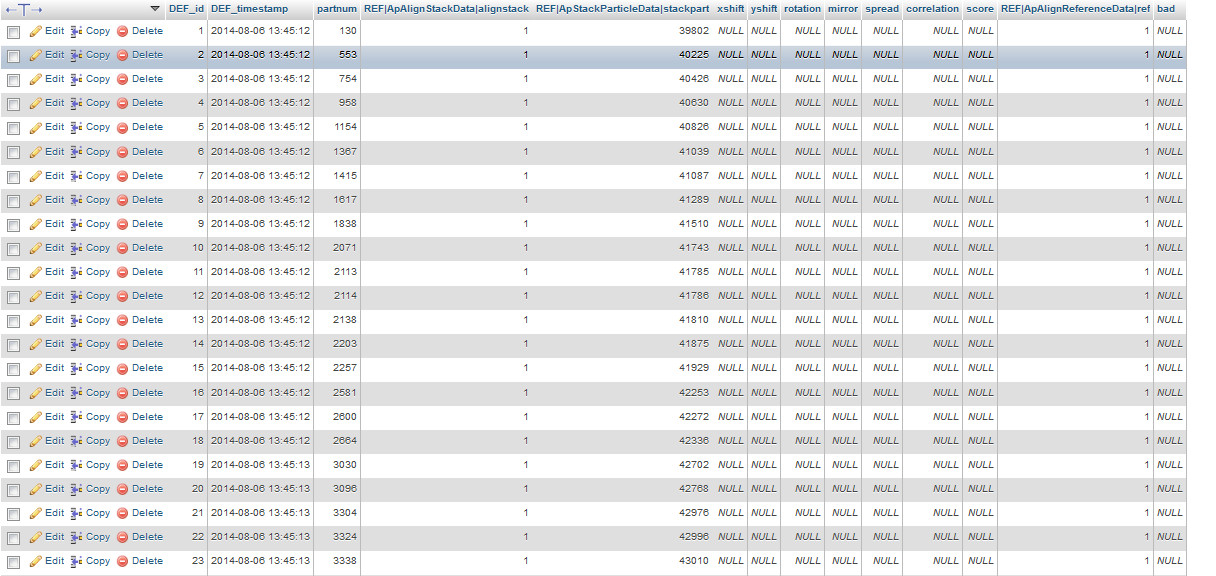
| ApAlignPartData.jpg (283 KB) ApAlignPartData.jpg | AplignPartData |
RE: rctvolume.py in appion 3.0 - Added by Neil Voss almost 11 years ago
Well, EMAN refine2d does NOT upload its alignment. As far as I know, Xmipp MaxLike, Xmipp Cluster2d, ISAC, and the SPIDER methods only upload the alignment parameters.
I am not really sure what the information I should be seeing in your images.
With the Xmipp MaxLike, it needs to have been uploaded (You should see the "Run Feature Analysis" button).
RE: rctvolume.py in appion 3.0 - Added by Jason van Rooyen almost 11 years ago
Sorry for the confusion. Xmipp maxlike (with upload) followed by kerden worked.
Ah, that explains it then. If it's not meant to upload then there isn't a problem (except for my expectation).
My understanding came from http://emg.nysbc.org/projects/appion/wiki/Random-Conical_Tilt_%28RCT%29_Reconstruction_workflow which I thought suggested that only cl2d did not upload.
Thanks for the help.
RE: rctvolume.py in appion 3.0 - Added by Neil Voss almost 11 years ago
So is it working for you now?
CL2D works with RCT in Xmipp 3.0, but not in Xmipp 2.4. I just found this out in August.
RE: rctvolume.py in appion 3.0 - Added by Jason van Rooyen almost 11 years ago
Yes, thanks Neil.
I got it working with maxlike followed by kerden self-organizing map.
I will try with cl2d after upgrading Xmipp as well.